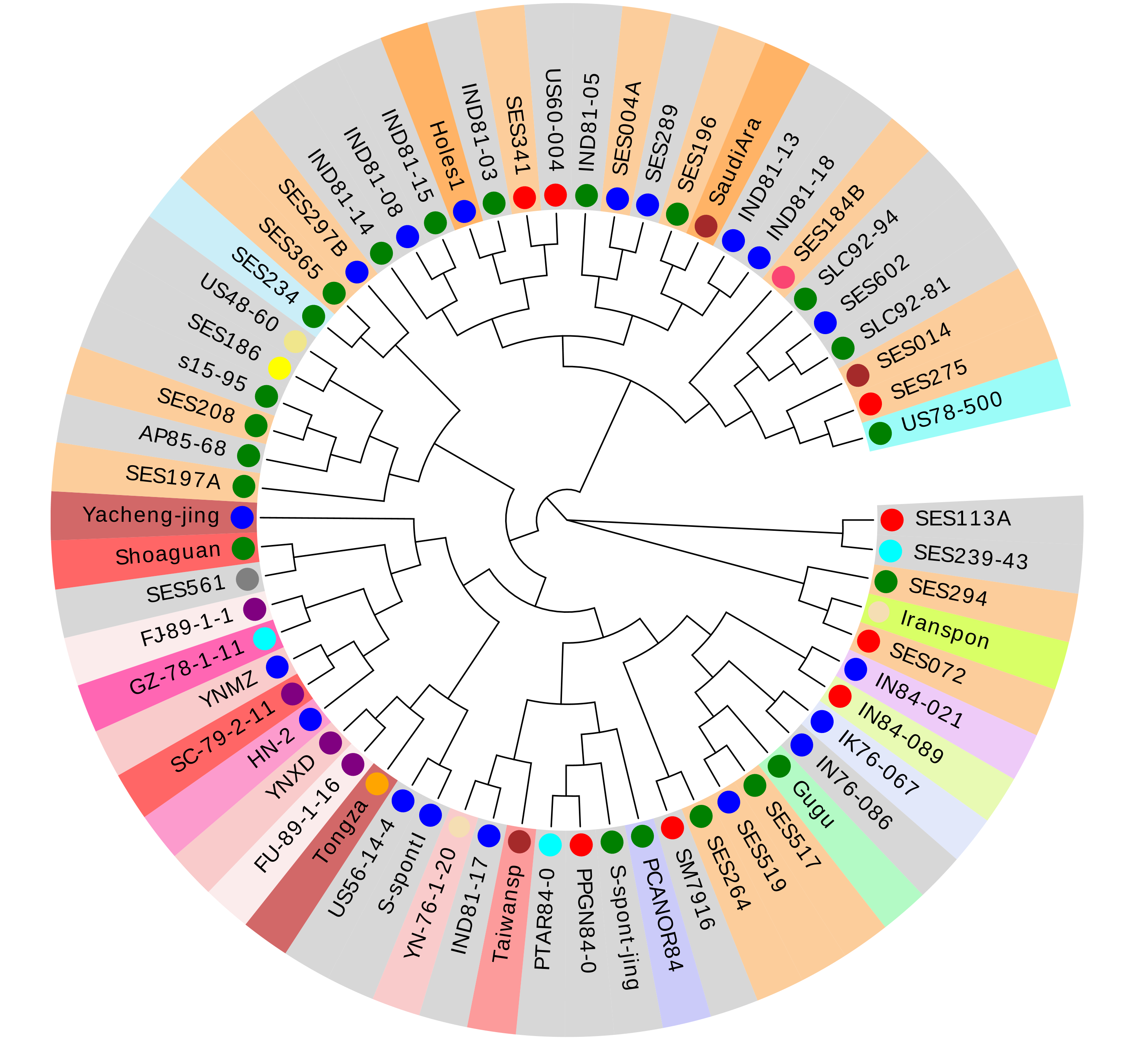
Single Nucleotide Polymorphism
The single nucleotide polymorphism (SNP) calling are based on a total of 64 genome resequencing of Saccharum accessions with the AP85-441 as reference genome by using BWA (Version: 0.7.12) with the default parameters. The mapping files were tranfer into the BAM format used SAMtools(Vesion:1.3) and the duplicated reads were filter by the Picard package.The SNP were detect by using the Genome Analysis Toolkit (GATK, version 3.7) . The neighbor-joining tree was constructed using SNPhylo software with the bootstrap 100. Finally,we identified 36,798,531 SNPs and 4,568,511 indels in the sugarcane genome. We implemented phylogenetic analysis of 64 accessions using FigTree the results are as follows.
Note:You may click here to download and known the varieties of information of every accession. you may observe the visualization of the SNP loci by JBrowse(Click on each mutation sites in JBrowse to check detail corresponding variability information of each accession)